Our Research
The purpose of computing is always to gain insights, not numbers (as Richard Hamming said). Today, there is a dire need to gain insights and observations much more efficiently than ever before, especially after witnessing both unprecedented growth in data volume and the start of the Angstrom era of semiconductors. Bioinformatics is one of the scientific disciplines that necessitate such efficient computation due to the complexity of data and problems that bioinformatics should solve timely. In bioinformatics, we develop methods and build architectures to solve biological problems. Understanding and analyzing biological data, such as genomes, is the foundation of many scientific and medical discoveries in biomedicine and other life sciences through its rapidly growing presence in clinical medicine, outbreak tracing, and understanding of pathogens and urban microbial communities.
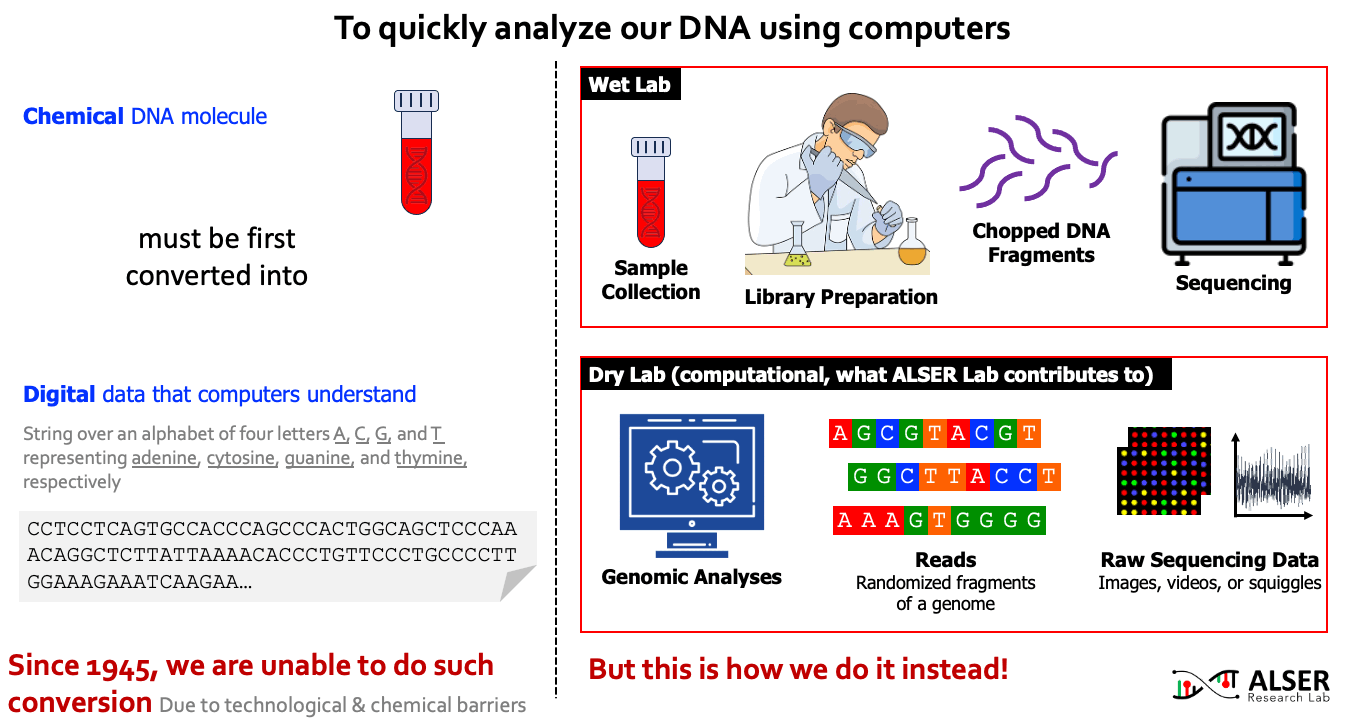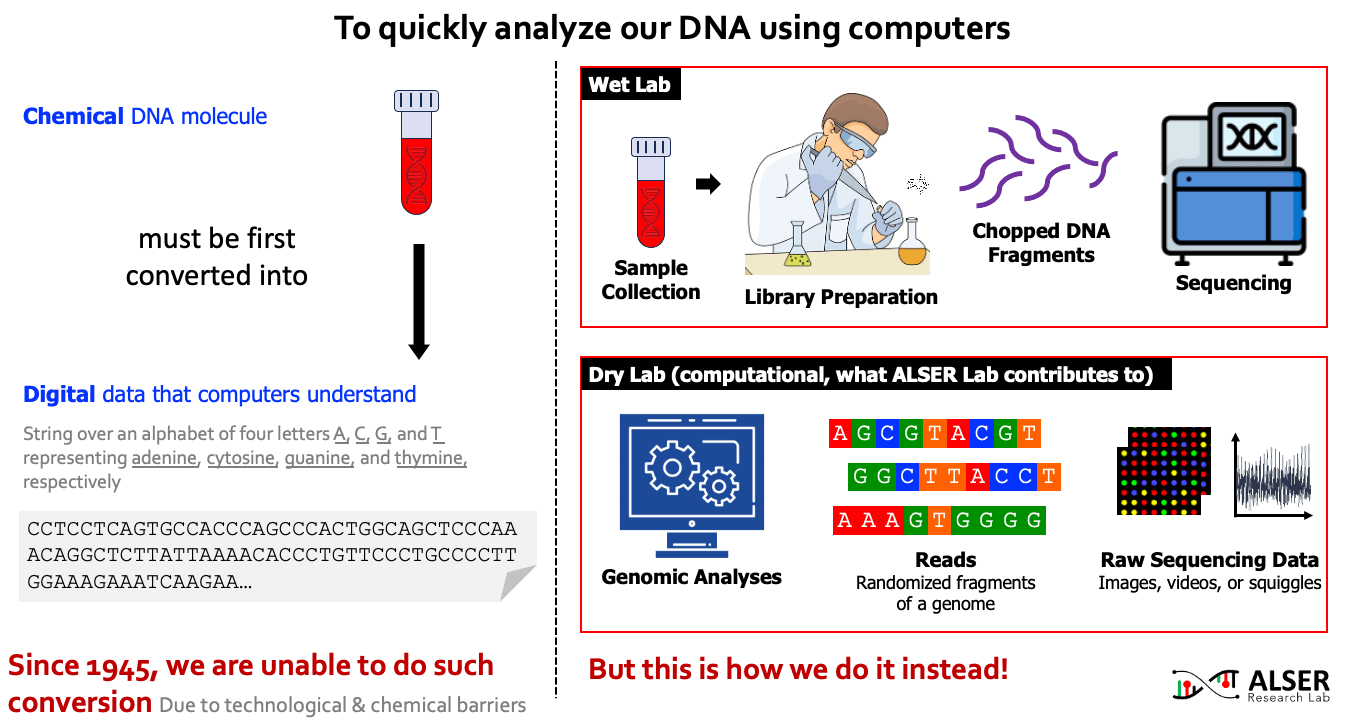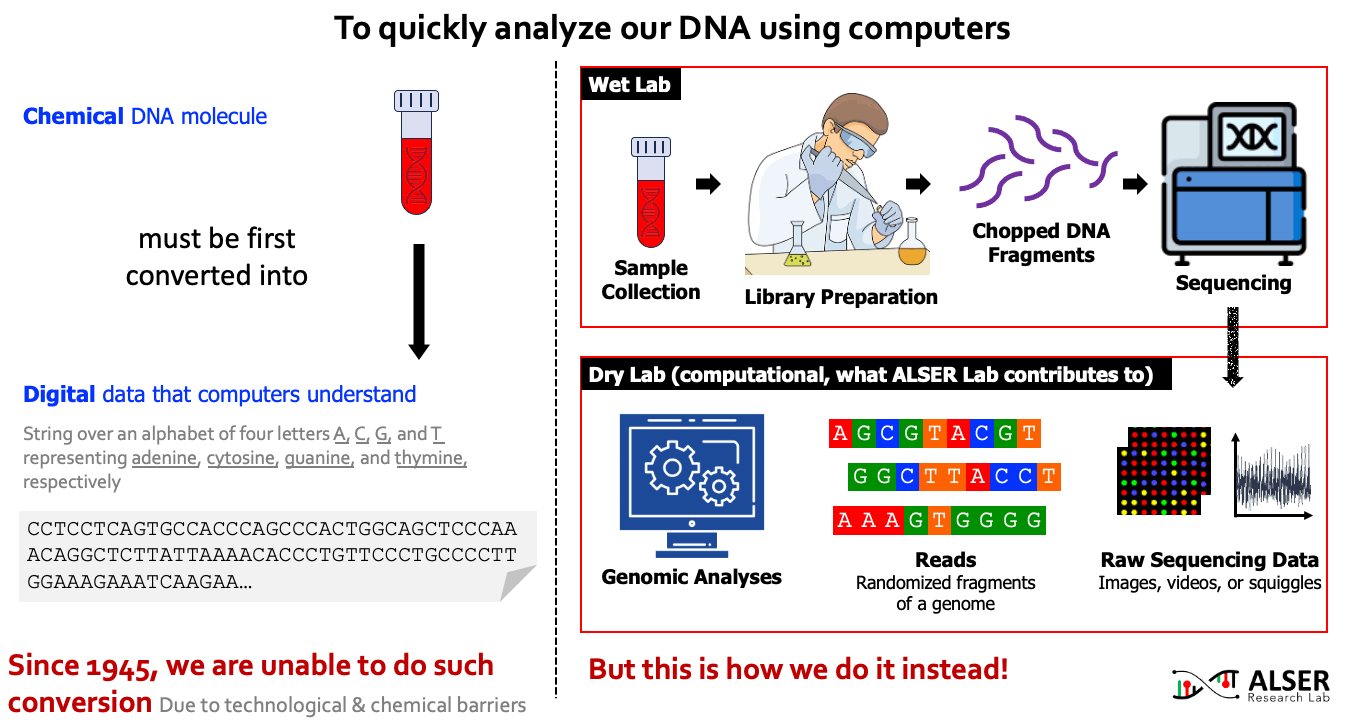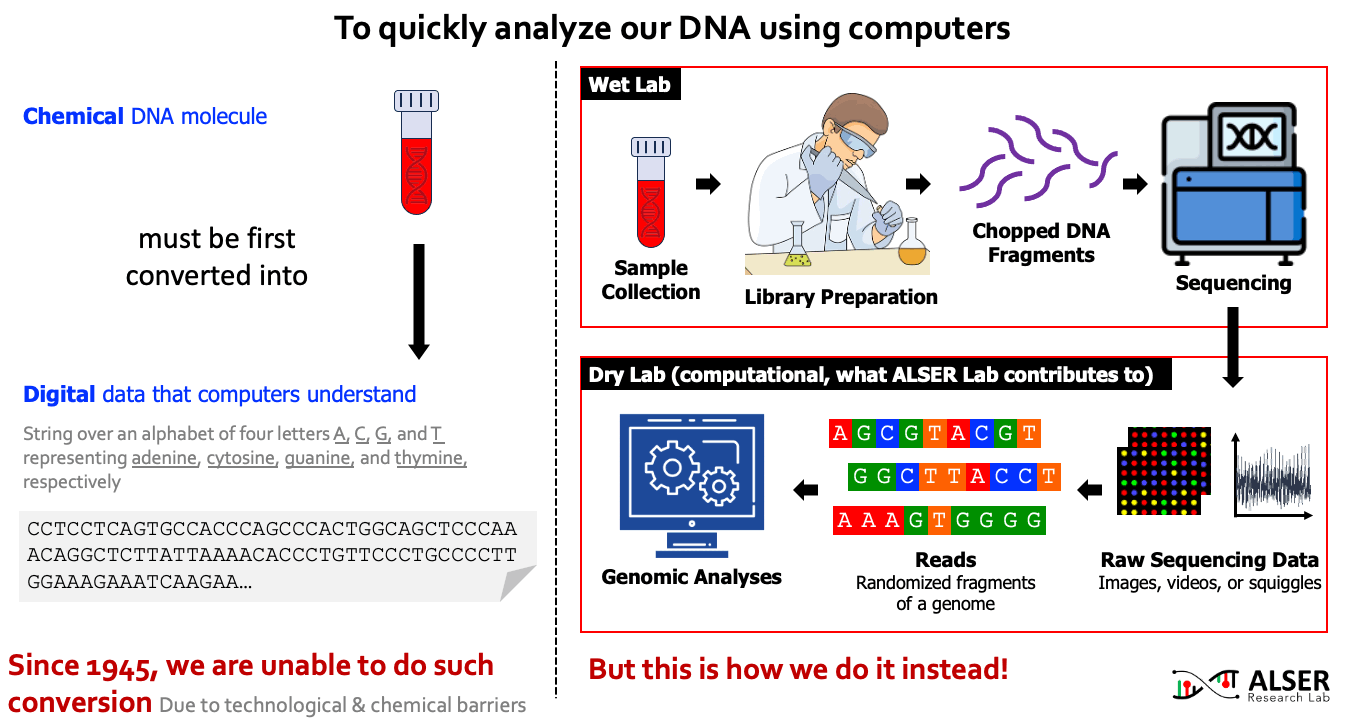
Our understanding of genomic molecules today is affected by the ability of modern computing technology to quickly and accurately determine an individual’s entire genome. To computationally analyze an organism’s genome, the DNA molecule must first be converted to digital data in the form of a string over an alphabet of four letters or base-pairs (bp), commonly denoted by A, C, G, and T. The four letters in the DNA alphabet correspond to four chemical bases, adenine, cytosine, guanine, and thymine, respectively, which make up a DNA molecule. After more than 7 decades of continuous attempts (since 1945, the first attempt to read the full content of Insulin protein and later was granted a Nobel prize), there is still no sequencing technology that can read a DNA molecule in its entirety. As a workaround, sequencing machines generate randomly sampled subsequences of the original genome sequence, called reads. The resulting reads lack information about their order and corresponding locations in the complete genome. Software tools, collectively known as genome analysis tools, are used to reassemble read fragments back into an entire genome sequence and infer genomic variations that make an individual genetically different from another.
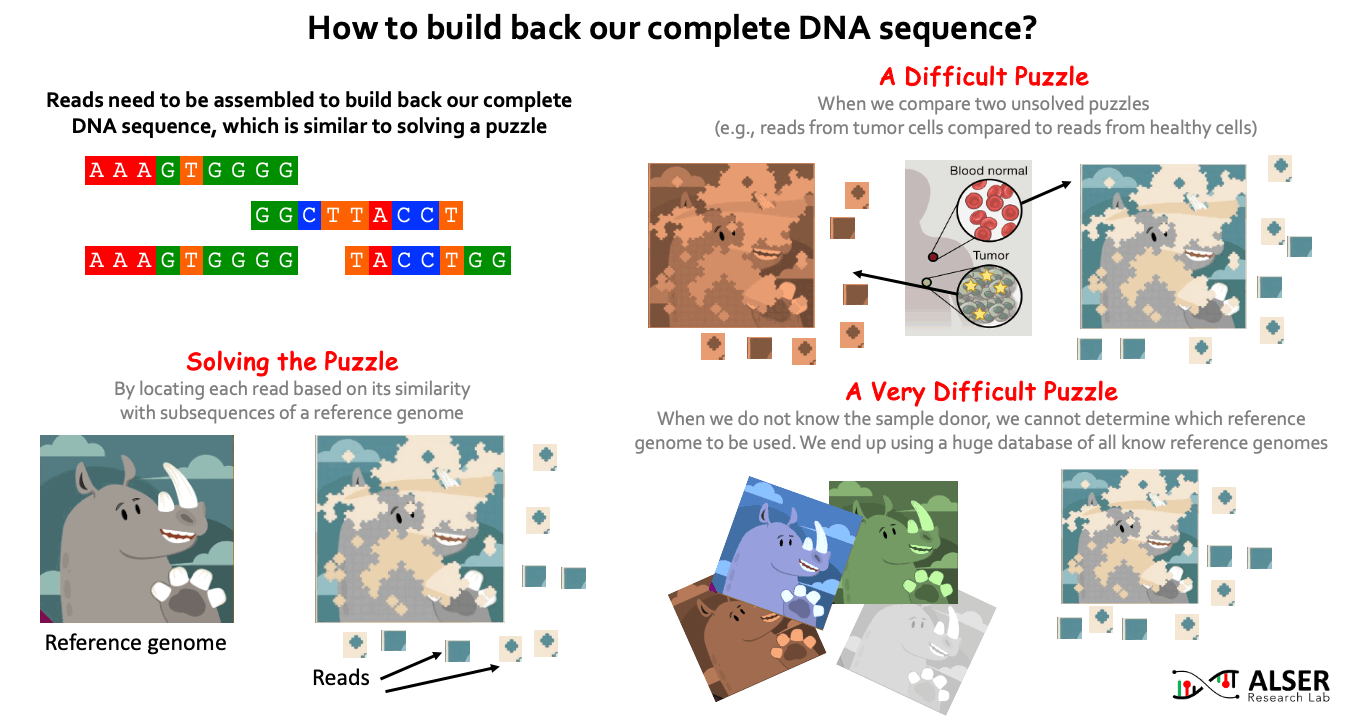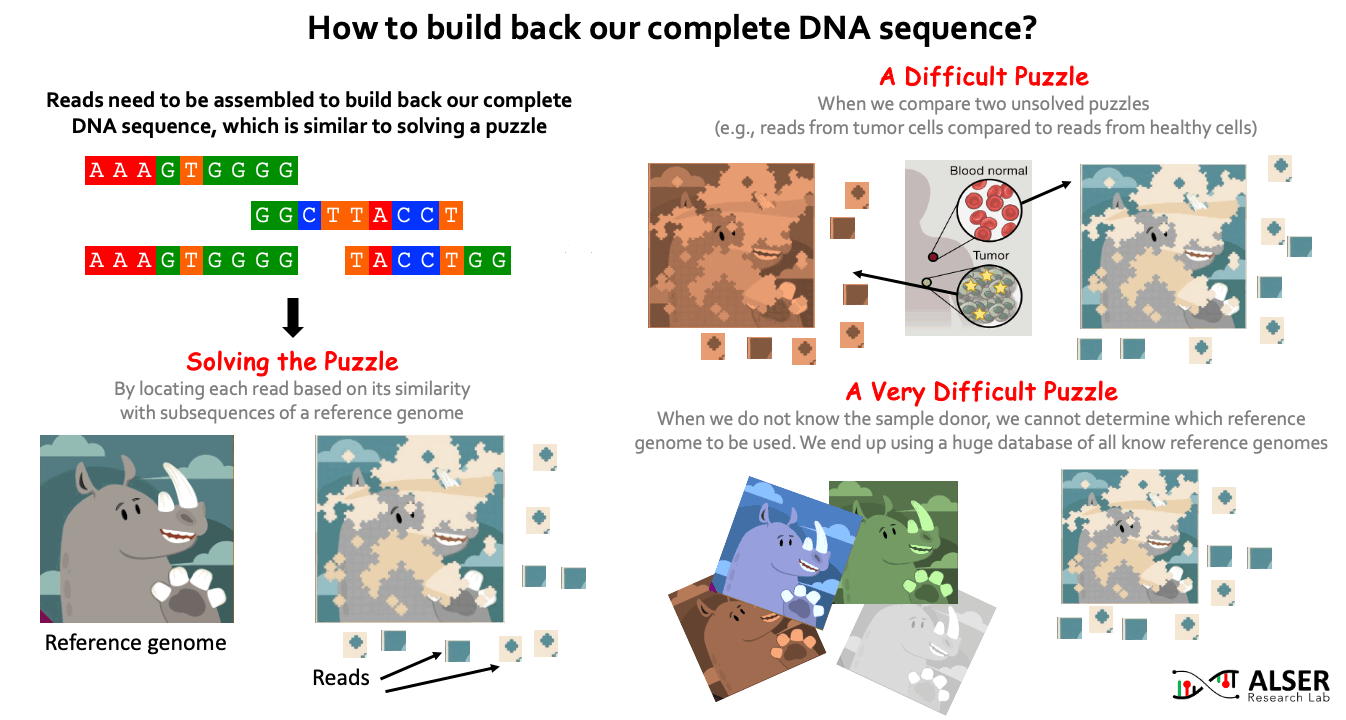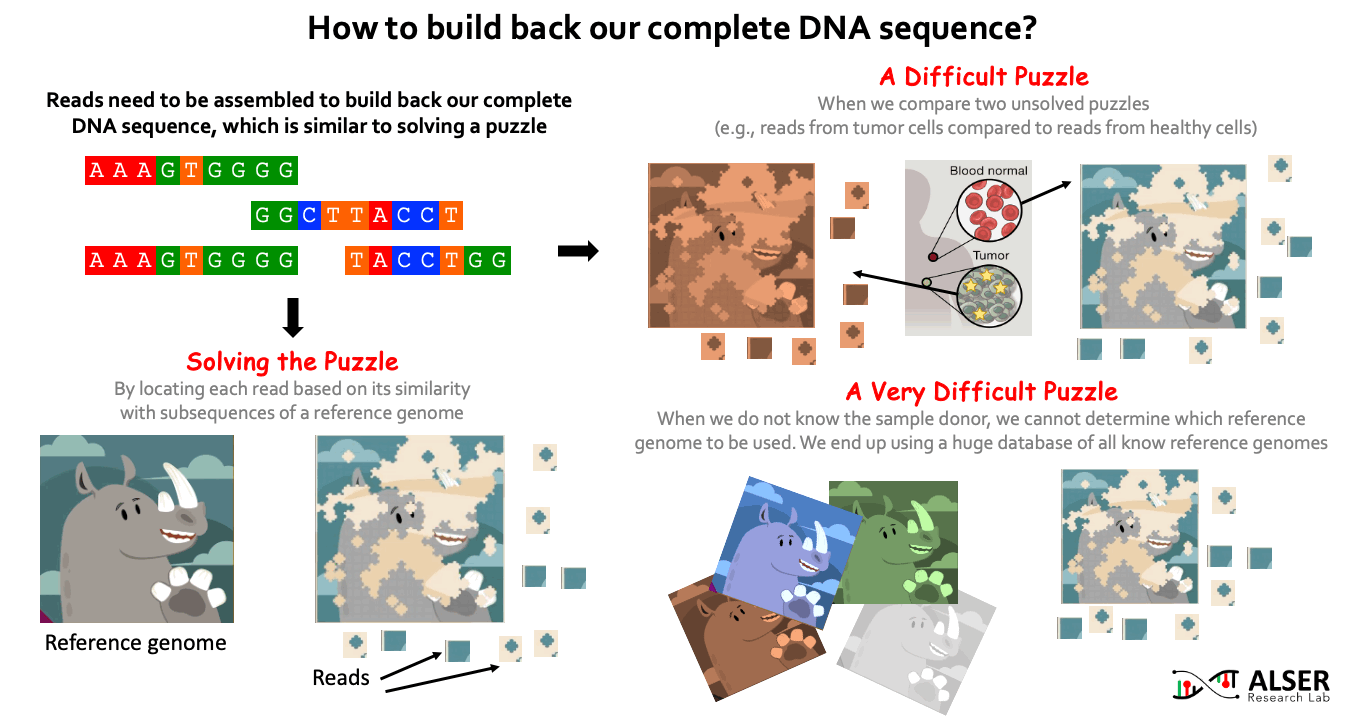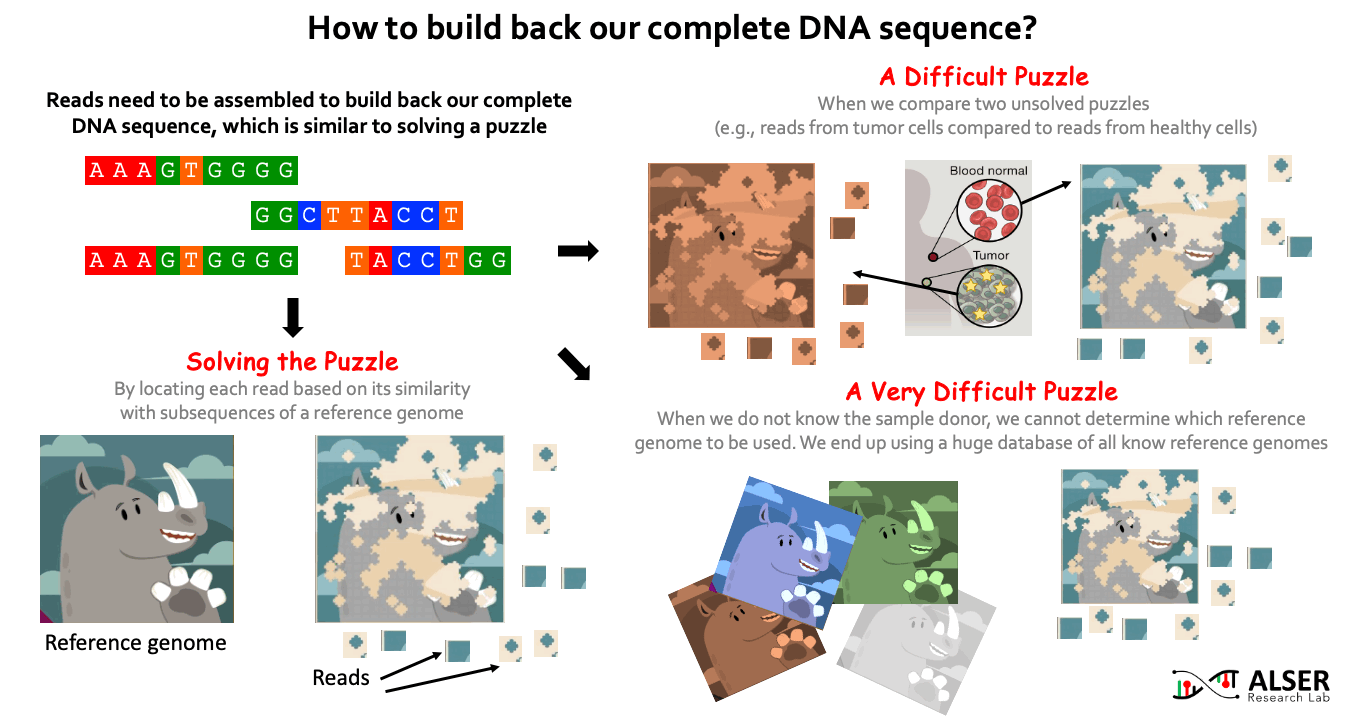
Since sequencing genomic data provides randomized fragments of the actual genome sequence, we need to reassemble back these fragments. Similar to solving jigsaw puzzles, we match each read sequence to one or more possible locations within a reference genome (i.e., a representative genome sequence for a particular species) based on the similarity between the read and the reference sequence segment at that location. When the sample donor is known, we can obtain a reference genome for the donor species from public reference databases (e.g., RefSeq, PATRIC, and Ensembl). It is more challenging when we aim to identify differences (somatic variations) between two sets of reads, for example, try to understand what genetic variations cause a cancer by comparing reads sequenced from normal and cancerous cells of the same person. It can be much more challenging when we do NOT know the sample donor. This requires comparing the reads to ALL known reference genomes (4 TB of data in RefSeq alone). For example, when collecting a genomic sample from a train seat, we expect to have a collection of bacterial and viral species (and probably others) present in the sample. For that, the detection of species in the sample highly depends on what species we have already known before and already stored in reference databases.
When we match each read sequence to each potential location within a reference genome, the bases in a read may, unfortunately, not be identical to the bases in the reference genome at the location that the read actually comes from. These differences may be due to (1) sequencing errors (with a rate in the range of 0.1–20% of the read length, depending on the sequencing technology), and (2) genetic differences that are specific to the individual organism’s DNA and may not exist in the reference genome. We must tolerate such differences during similarity checks, which makes it even more challenging. When we build back the sequenced genome, we aim to identify the possible genetic differences between the reference genome and the sequenced genome. Genetic differences include small variations that are less than 50 bp, such as single nucleotide polymorphisms (SNPs) and small insertions and deletions (indels). Genetic differences can also be larger than 50 bp variations, known as structural variations, which are caused by chromosome-scale changes in a genome. For example, the insertion of about 600,000-base long region has been observed in some chromosomes.
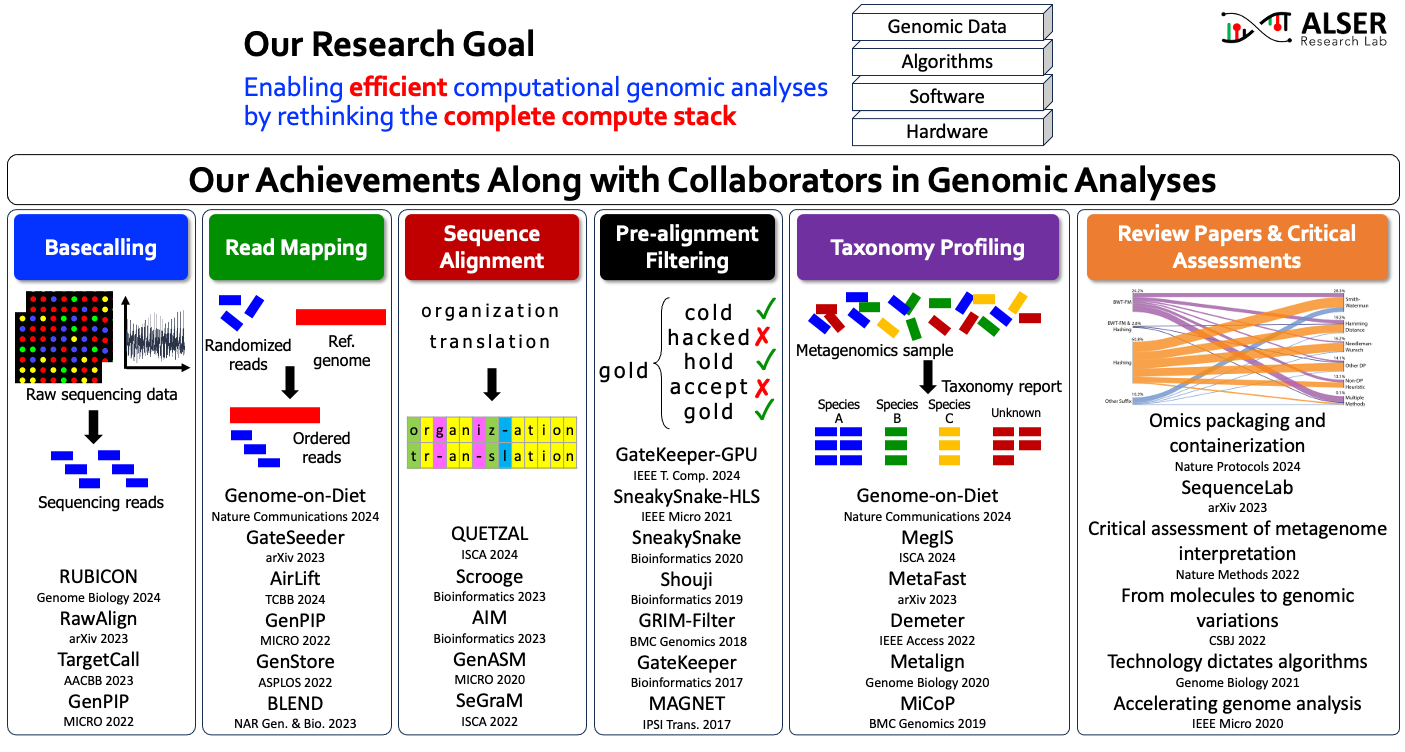
We developed new algorithms and architected different solutions and designs that are now an integral part of genomic pipelines and used to solve challenging biological problems. As most bioinformatics applications are implemented as computer programs, there is always an urgent and constant need to improve bioinformatics software tools that can benefit significantly from algorithmic optimizations and adapting new computing techniques. We believe that rethinking existing bioinformatics pipelines to redefine roles played by each computational step can provide transformational benefits. Our long-term goal is to develop and enable truly intelligent genomic analyses, where genomic data are analyzed efficiently at a truly population scale. The lag between data generation and obtaining analysis results may pose problems in rapid clinical diagnosis, for example, in identifying infectious agents, choosing the correct antibiotics for sepsis and hospital infections, identifying genetic disorders in critically ill infants, and tracking outbreaks that spread quickly in the population as evidenced by recent outbreaks. The problem is exacerbated in developing countries and during crises where access to data centers is even more limited. We are thrilled to continue improving bioinformatics applications, especially those that are essential components of biomedical and clinical applications. We are eager to collaborate with other scientists to achieve practical and realizable complete solutions using existing and emerging technologies, such as near-data processing, quantum computing, neuromorphic computing, specialized architectures, and reconfigurable fabrics. Looking forward, we are excited about working with medical domain experts to identify and optimize other challenging clinical applications that can benefit from my expertise in performance improvement.