
FASTR: Reimagining FASTQ via Compact Image-Inspired Representation
Authors
Adrian Tkachenko, Sepehr Salem, Ayotomiwa Ezekiel Adeniyi, Zülal Bingöl, Mohammed Nayeem Uddin, Akshat Prasanna, Alexander Zelikovsky, Serghei Mangul, Can Alkan, Mohammed Alser
ALSER Lab, Georgia State University · Bilkent University · Sage Bionetworks
Abstract
Motivation: High-throughput sequencing (HTS) enables population-scale genomics but generates massive datasets, creating bottlenecks in storage, transfer, and analysis. FASTQ, the standard format for over two decades, stores one byte per base and one byte per quality score, leading to inefficient I/O, high storage costs, and redundancy. Existing compression tools can mitigate some issues, but often introduce costly decompression or complex dependency issues.
Results: We introduce FASTR, a lossless, computation-native successor to FASTQ that encodes each nucleotide together with its base quality score into a single 8-bit value. FASTR reduces file size by at least 2x while remaining fully reversible and directly usable for downstream analyses. Applying general-purpose compression tools on FASTR consistently yields higher compression ratios, 2.47, 3.64, and 4.8x faster compression, and 2.34, 1.96, 1.75x faster decompression than on FASTQ across Illumina, HiFi, and ONT reads. FASTR is machine-learning-ready, allowing reads to be consumed directly as numerical vectors or image-like representations. We provide a highly parallel software ecosystem for FASTQ/FASTR conversion and show that FASTR integrates with existing tools, such as minimap2, with minimal interface changes and no performance overhead. By eliminating decompression costs and reducing data movement, FASTR lays the foundation for scalable genomics analyses and real-time sequencing workflows.
Features
- FASTR is at least 2x less in size than FASTQ, and hence better to read, process, transfer
- FASTR can be further compressed using general-purpose compression tools, such as gzip, pigz, ...
- Extremely fast (multithreaded) and lossless FASTR-to-FASTQ & FASTQ-to-FASTR conversion
- FASTR supports data from all prominent sequencing technologies (Illumina, ONT, PacBio's HiFi, and PacBio's CLR), single-end and paired-end reads, and SRA formats (https://www.ncbi.nlm.nih.gov/sra)
- FASTR supports all widely-used Phred quality scores (Phred42, Phred63, Phred68Solexa, Phred94, Illumina RTA3, Illumina RTA4, and custom mathematical formulas)
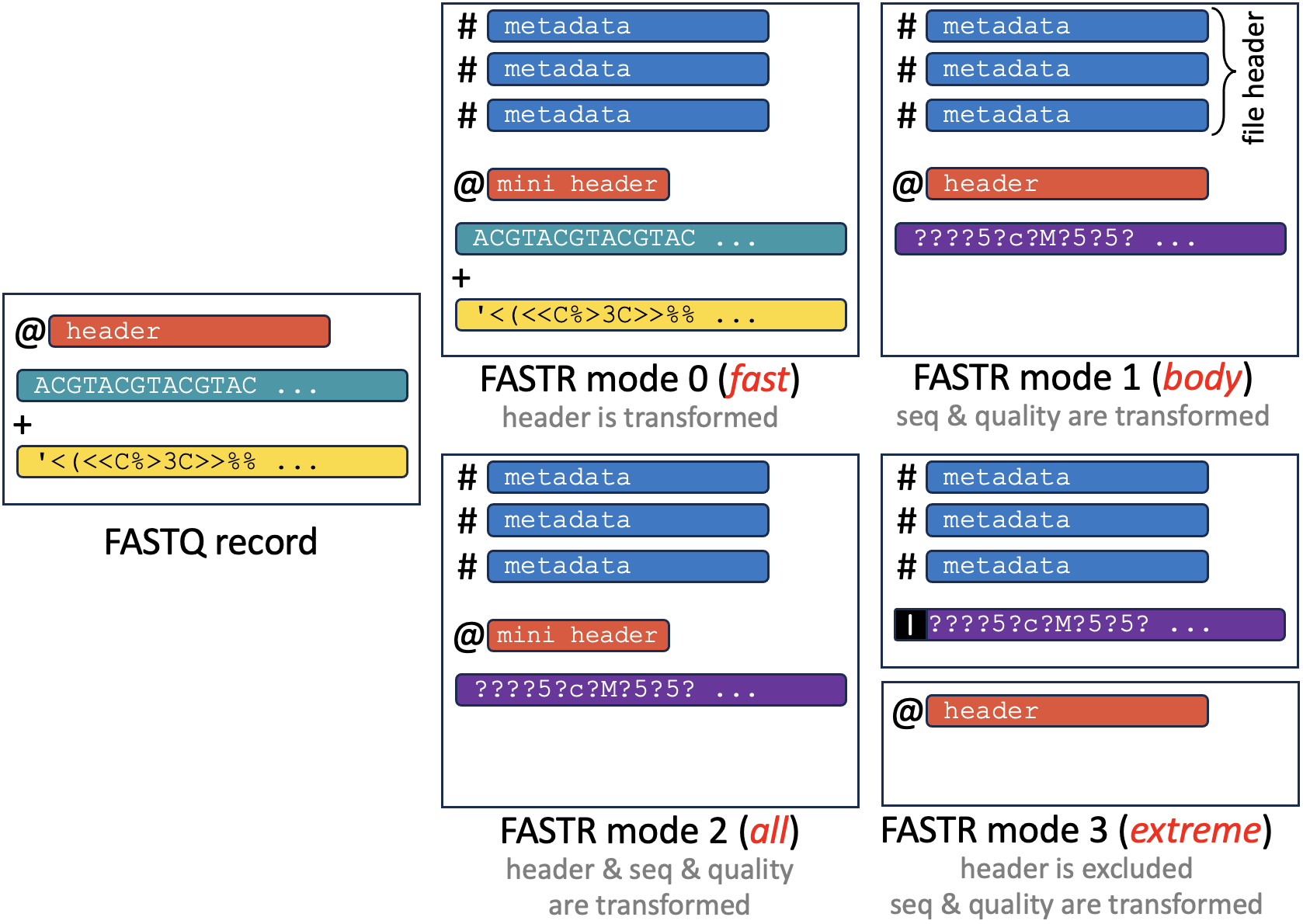
- Flexible Output: binary (1 uint8 per FASTR value), integer (3 uint8s per FASTR value), with/without header
- FASTR is compatible with minimap2 with no (or <2%) overhead, and with machine learning pipelines (i.e., numerical vectors)
Availability
- GitHub: https://github.com/ALSER-Lab/FASTR
- BioRxiv: https://www.biorxiv.org/content/10.64898/2026.01.22.701172v1
- Visualizer: https://alserlab.github.io/tools/fastr/FASTR_Visualizer/
Keywords
FASTQ, FASTR, genomics, sequencing, compression, machine learning, high-throughput sequencing